MOLDIMED – HPC-ready analytical pipeline solution for DNA massive parallel sequencing
INTRODUCTION:
Nowadays, massive parallel sequencing (MPS) have started to play a key role in DNA diagnostics. MPS methods generate enormous amount of data and require command line based analytical tools linked to Linux, what is often limiting factor for potential users. Here we demonstrate straight-forward and user-friendly solution for MPS variant analysis.
TECHNOLOGY (INVENTION) DESCRIPTION:
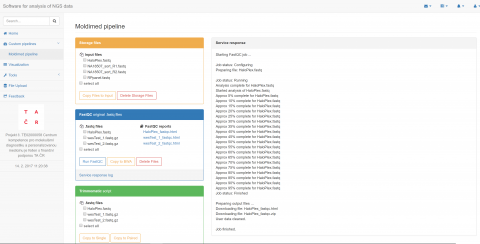
Designed pipeline is separated into two main parts. The first part contains processing of raw sequencing reads including quality check, read trimming, alignment to reference genome and variant calling. Default settings is based on general recommendations of good practice, nevertheless attributes might be configurable by a user. The computational steps are carried out by integration of several GNU licensed tools. The second part provides annotation of obtained DNA variants, their localization on gene level and effect on translation. Variants are characterized by frequency in population, pathogenicity, damaging protein predictions in silico and linked to gene ontologies and human phenotype ontologies terms.
ADVANTAGES OVER EXISTING SOLUTIONS:
The compact and intuitive platform enabling analysis from raw data to variants annotation. Variant filtering options are allowed by frequency, pathogenicity, diseases phenotype or selected molecular pathways criteria. Moreover, variant characterization could be extended by uploading of user’s custom files in standard formats (VCF/GTF/GFF). Besides the human genome, annotation software has been successfully tested for variants in non-model and incomplete genomes such as barley and some fungi. The goal of the platform is also to provide easy and intuitive access to HPC computing resources to scientific researchers in the area of clinical MPS data.
DEVELOPMENT STATUS (STAGE):
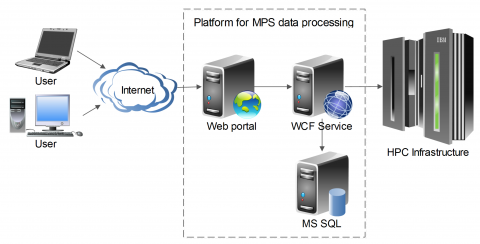
The first version of the platform is developed and currently using IT4I’s supercomputers Anselm and Salomon.
PUBLICATIONS:
IP PROTECTION STATUS:
Registered software (RIV) – Web GUI for high capacity DNA sequencing tools and human genome DNA variants annotation.
TECHNOLOGY / IP OWNERS :
Václav Svatoň, IT4Inovations, VŠB-TU Ostrava Petr Vojta, IMTM, Univerzita Palackého Olomouc Marián Hajdúch, IMTM, Univerzita Palackého Olomouc Zuzana Macečková, IMTM, Univerzita Palackého Olomouc
More information
More information is available upon signing a CDA / NDA (Confidential Disclosure Agreement / Non-Disclosure Agreement)