Software system for reliable identification of mutant and polymorphic proteins from mass spectrometric data
INTRODUCTION:
Mutational profiling of biological systems is highly informative as it sheds light on altered processes and guides selection of therapies. Although regularly performed on DNA/RNA level at present, recent developments enable use of mass spectrometry for identification of mutant proteins with corresponding applications in personalized medicine.
TECHNOLOGY (INVENTION) DESCRIPTION:
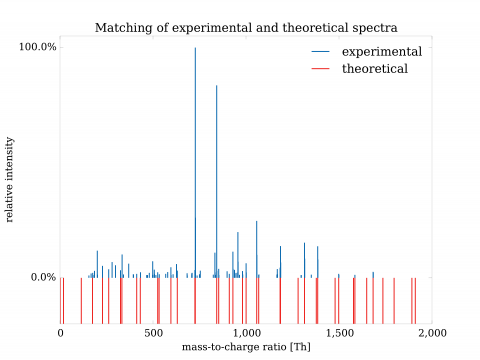
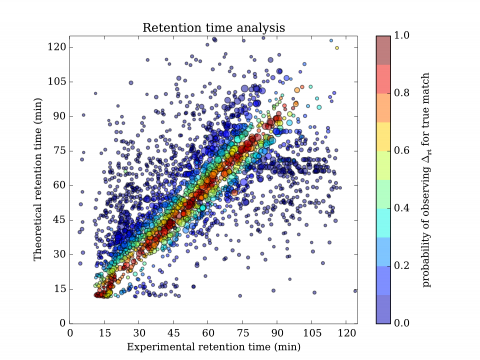
In general, identification of mutations from mass spectrometric data of complex sample poses significant challenges and with usual approach yields high rate of false findings. The problem is mainly due to large size of similarly-viable interpretations of measured spectra. To better understand the situation, we have developed both a mathematical theory of identification and a system which implements it. With incorporation of additional information, it is then possible to dramatically restructure the possible interpretations and often pinpoint particular interpretation as the optimal one. The approach was used for identification of altered proteins and exhibited precise, selective elimination of false results. The technology could be used to reliably identify mutant and polymorphic proteins.
ADVANTAGES OVER EXISTING SOLUTIONS:
The advantages could be compared to a) nucleotide-based solutions such as DNA/RNA sequencing or b) other solutions in proteomics. In the former case, the approach has fundamental advantage of direct observation of expressed proteins; their altered and unaltered forms, quantities and effects of mutations on presence of relevant post-translational modifications. In the latter case, the solutions either do not exist at all or are partial and lacking large-scale validation. Validations done in our case turned out to be crucial and showed need for extensive development of both mathematical and computational aspects of identification to limit otherwise unacceptable rate of false positives.
DEVELOPMENT STATUS (STAGE):
Validation on a wide range of internal and publicly available data (≈ 6 TB). Software accessible through web interface.
PUBLICATIONS:
IP PROTECTION STATUS:
TECHNOLOGY / IP OWNERS :
Palacky University Olomouc — Institute of Molecular and Translational Medicine (IMTM), Faculty of Medicine and Dentistry.
More information
More information is available upon signing a CDA / NDA (Confidential Disclosure Agreement / Non-Disclosure Agreement)